Overview of DNA Replication
Overview of DNA Replication
DNA replication is the process in which new copy of DNA is produced from parent DNA. When two strand of DNA are separated, each strand act as template for the formation of new strand. This process is called DNA replication.
Mechanism of DNA Replication
Steps of DNA replication are:
- Initiation
- Chain Elongation
- Termination
1. Initiation
- Replication starts at a specific point of DNA called origin of chromosomal replication (Ori C). In prokaryotic cell, there is one Ori whereas in Eukaryotic cell number of Ori is more than one.
- Ori C is a 245 base pair region of the chromosome and bears DNA sequence elements i.e. 2 types of repeated sequence.
i. Four 9 mer motif which is the binding site for DnaA.
ii. Three 13 mer morif which is the initial site of single stranded DNA formation. - There is a formation of replication bubble due to the presence of origin of replication.
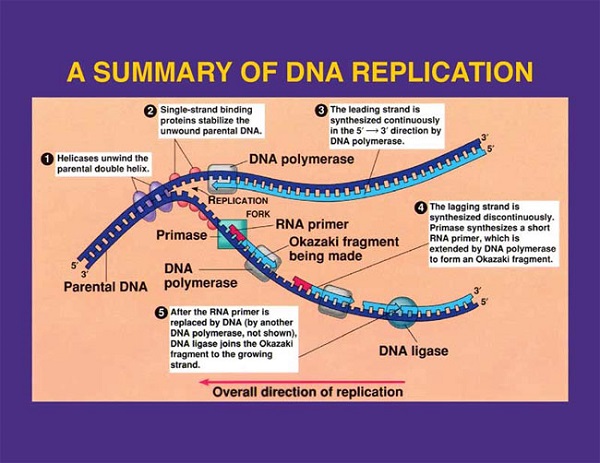
- At first, double stranded DNA starts to separate and uncoil due to breaking of hydrogen bond present between nitrogen bases by helicase. The unwinding DNA molecule takes place once in every ten nucleotide pair in eukaryotic DNA.
- Each uncoiled parental DNA strand acts as template DNA strand for the synthesis of new complementary strands.
- When two strands unwind and separate incompletely, they form Y-shape where active synthesis occurs. This region is called replication fork.
- Each separated strands are stabilized by single stranded binding protein.
- As the two strands are separated, supercoiling occurs which is removed by DNA topoisomerase.
- Initiation of replication requires RNA primer, which is a small strand of RNA, synthesized by primase.
2. Chain Elongation
- Chain elongation proceeds from the initiation site by the addition of deoxyribonucleotides at 3’-OH end of the primer by DNA polymerase III.
- DNA Polymerase III forms continuous strand of DNA on 3’→5’ template. The continuous strand of DNA is called leading strand. Since the direction of movement of replication fork and direction of leading strand synthesis are same, leading strand is synthesized continuously after its initiation.
- However, in other template strand 5’→3’, there is discontinuous formation of DNA and thus more RNA primer are required for the formation of whole strand. Due to discontinuous formation, smaller fragments are formed, which are called Okazaki fragments. DNA ligase joins these Okazaki fragments to form complete lagging strands. Since the direction of movement of replication fork is opposite to direction of lagging strand synthesis, it cannot be synthesized continuously.
- After completion of chain elongation RNA primer is removed by exonuclease activity of DNA polymerase I and the gap is filled with complementary bases.
3. Termination
- Replication must be terminated to produce two daughter DNA molecule and to regulate and co-ordinate replication with cell division.
- When two replication fork meets Ter-Tus complex, DNA synthesis stops. And the daughter DNA are produced.
- In bacteria DNA is circular. Therefore two interlinked daughter DNA are obtained at completion of replication. Such interlinked DNA are called catenanes. Finally DNA topoisomerase IV cuts one DNA, removes it out of other and finally reseals it. So that two daughter DNA are separated. This process is known as decatenation.

I love this site bro…..wonderful
thank you for putting notes
Weldon bro good and thanks for sharing this articlle